Code
library(tree)
library(ISLR2)Back to basics
March 2, 2023
All the code here is derived from the legendary book ISRL 2nd edition’s chapter 8 “Decision Trees”. Its sometimes a wonder how elegant the base R language can be. The ISRL lab rarely mentions tidyverse syntax but yet manages to make the code so easy to read. The more you learn!🤓
In today’s lab, we will be using the Carseats dataset from the ISLR2 package.
'data.frame': 400 obs. of 11 variables:
$ Sales : num 9.5 11.22 10.06 7.4 4.15 ...
$ CompPrice : num 138 111 113 117 141 124 115 136 132 132 ...
$ Income : num 73 48 35 100 64 113 105 81 110 113 ...
$ Advertising: num 11 16 10 4 3 13 0 15 0 0 ...
$ Population : num 276 260 269 466 340 501 45 425 108 131 ...
$ Price : num 120 83 80 97 128 72 108 120 124 124 ...
$ ShelveLoc : Factor w/ 3 levels "Bad","Good","Medium": 1 2 3 3 1 1 3 2 3 3 ...
$ Age : num 42 65 59 55 38 78 71 67 76 76 ...
$ Education : num 17 10 12 14 13 16 15 10 10 17 ...
$ Urban : Factor w/ 2 levels "No","Yes": 2 2 2 2 2 1 2 2 1 1 ...
$ US : Factor w/ 2 levels "No","Yes": 2 2 2 2 1 2 1 2 1 2 ...Creating a column called High which takes a Y/N value depending on the sales and then merge it with the Carseats df.
CarseatsCreating a classification tree to predict High using all variables except Sales
Classification tree:
tree(formula = High ~ . - Sales, data = Carseats)
Variables actually used in tree construction:
[1] "ShelveLoc" "Price" "Income" "CompPrice" "Population"
[6] "Advertising" "Age" "US"
Number of terminal nodes: 27
Residual mean deviance: 0.4575 = 170.7 / 373
Misclassification error rate: 0.09 = 36 / 400 Misclassification error of 9% is a good fit. Let’s try plotting it
checking the top few rows of predicted columns
[1] Yes No No Yes No No
Levels: No YesComparing predicted with actual values
What’s the accuracy?
77% Accuracy
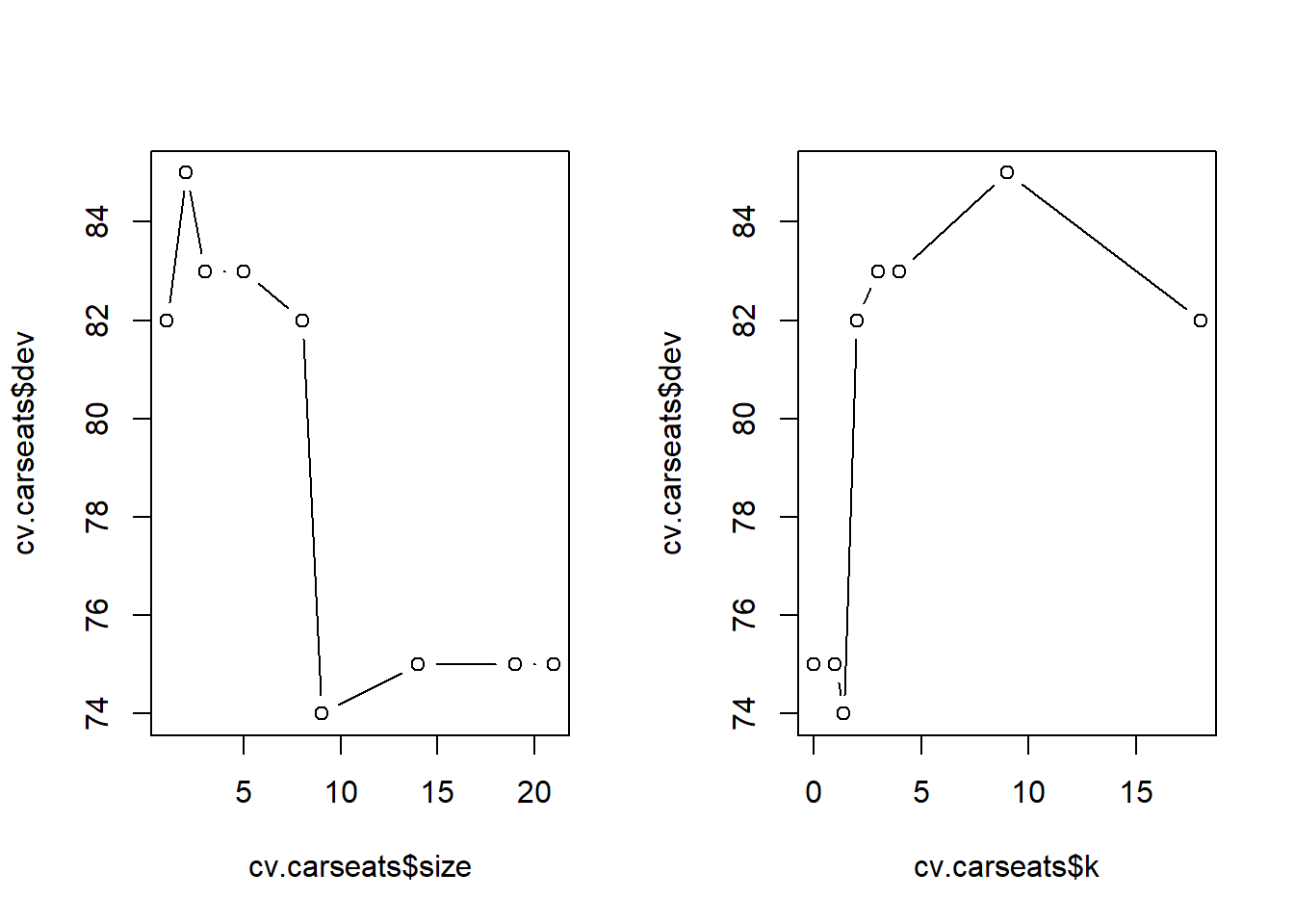
To improve the accuracy, lets attempt to prune the tree. For this cv.tree() function is used to determine the optimal level of tree complexity. Here the FUN argument is taken as prune.misclass to indicate that the cross-validation and tree pruning should be guided by the classification error instead of the default deviance.
[1] "size" "dev" "k" "method"Note to self:
k is the regularisation parameter size is # of terminal nodes for each treedev is the number of cross-validation errors$size
[1] 21 19 14 9 8 5 3 2 1
$dev
[1] 75 75 75 74 82 83 83 85 82
$k
[1] -Inf 0.0 1.0 1.4 2.0 3.0 4.0 9.0 18.0
$method
[1] "misclass"
attr(,"class")
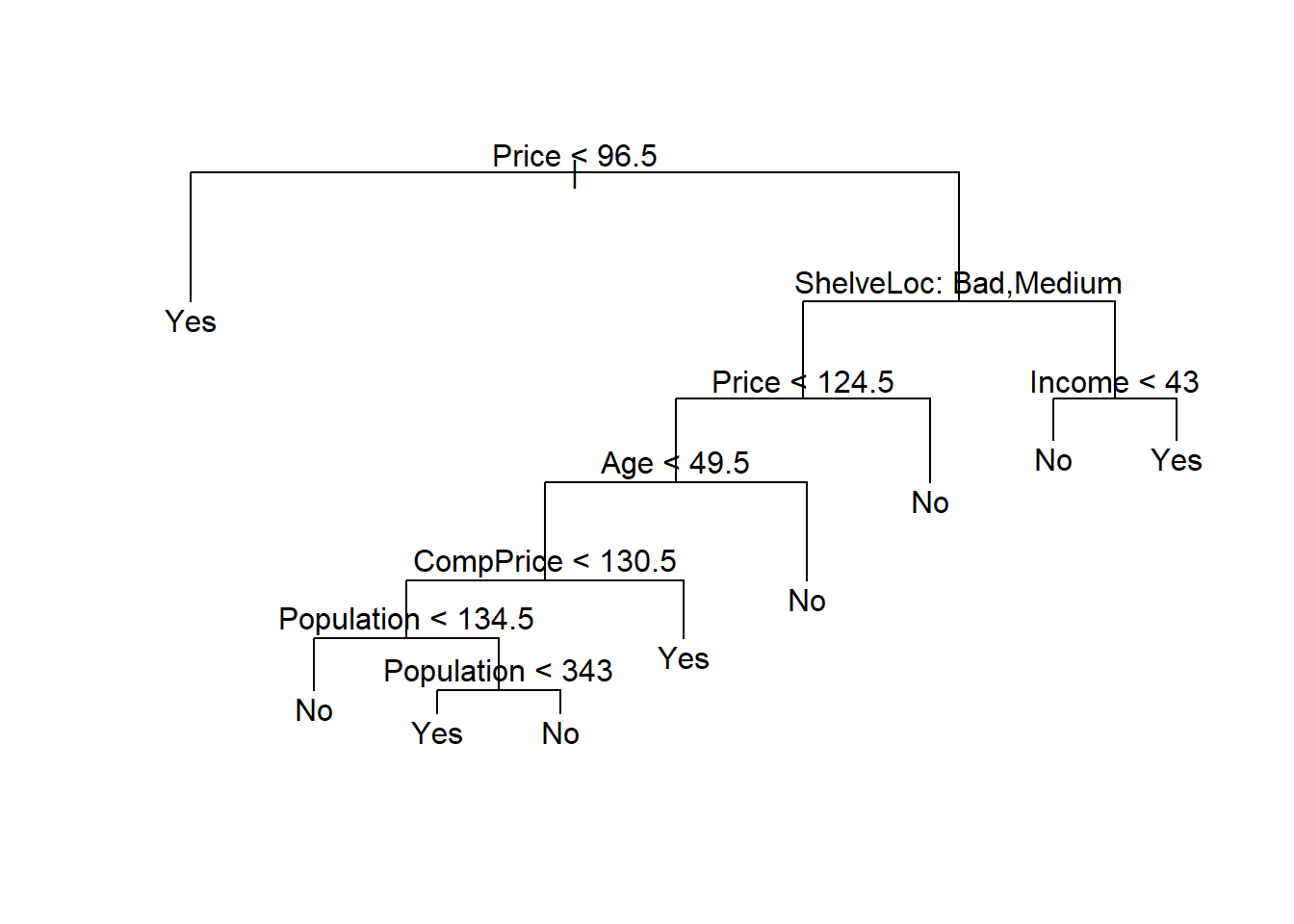
[1] "prune" "tree.sequence"Visualising the tree. The classification error is least (74) at size = 9
Using the prune.misclass() function to prune the tree to the 9-node specification.
Checking the accuracy in the good-old fashioned way (its really that simple!)🤓
High.test
prune.tree.pred No Yes
No 97 25
Yes 20 58So what’s the accuracy?
77.5% which is slightly better than the non-pruned tree. Not bad.
tidymodels interface allows for managing the resulting output and models in a more structured way.Boston datasetBoston dataset contains housing values of 506 suburbs of Boston. We are trying to predict the median value of the owner-occupied homes medv
'data.frame': 506 obs. of 13 variables:
$ crim : num 0.00632 0.02731 0.02729 0.03237 0.06905 ...
$ zn : num 18 0 0 0 0 0 12.5 12.5 12.5 12.5 ...
$ indus : num 2.31 7.07 7.07 2.18 2.18 2.18 7.87 7.87 7.87 7.87 ...
$ chas : int 0 0 0 0 0 0 0 0 0 0 ...
$ nox : num 0.538 0.469 0.469 0.458 0.458 0.458 0.524 0.524 0.524 0.524 ...
$ rm : num 6.58 6.42 7.18 7 7.15 ...
$ age : num 65.2 78.9 61.1 45.8 54.2 58.7 66.6 96.1 100 85.9 ...
$ dis : num 4.09 4.97 4.97 6.06 6.06 ...
$ rad : int 1 2 2 3 3 3 5 5 5 5 ...
$ tax : num 296 242 242 222 222 222 311 311 311 311 ...
$ ptratio: num 15.3 17.8 17.8 18.7 18.7 18.7 15.2 15.2 15.2 15.2 ...
$ lstat : num 4.98 9.14 4.03 2.94 5.33 ...
$ medv : num 24 21.6 34.7 33.4 36.2 28.7 22.9 27.1 16.5 18.9 ...Creating the training set for Boston which is half the size of the original
Building the tree
Regression tree:
tree(formula = medv ~ ., data = Boston, subset = train.boston)
Variables actually used in tree construction:
[1] "rm" "lstat" "crim" "age"
Number of terminal nodes: 7
Residual mean deviance: 10.38 = 2555 / 246
Distribution of residuals:
Min. 1st Qu. Median Mean 3rd Qu. Max.
-10.1800 -1.7770 -0.1775 0.0000 1.9230 16.5800 only 4 predictors rm, lstat, crim, age were used. (wonder why?) Plotting the decision tree
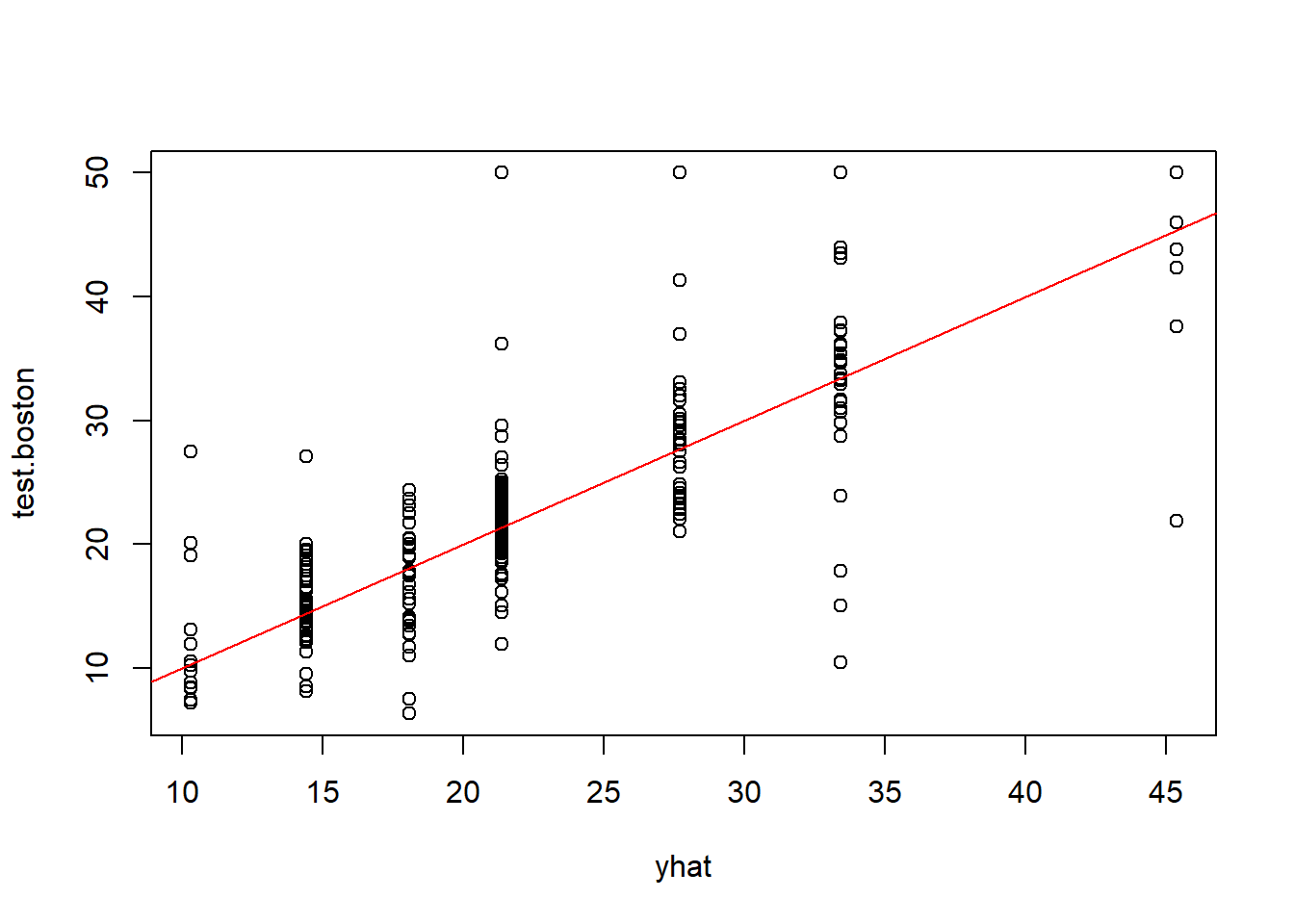
Mean Square Error is defined as
RMSE which uses the same units as the output variable is:
As the SD is the same units as the outcome variable, we can say that this model leads to predictions which on an average are within ±$5940 of the true median home value. Can we do better? Let’s keep digging
Note: Bagging is a special case of Random Forest where randomForest() function can be used for evaluating predictions from both bagging & RF. So first up is the Bagging process
importance parameter here will compute and return the importance measures of each predictor variable. Importance measures provide a way to assess the relative importance of each predictor variable in the random forest model, based on the decrease in accuracy that occurs when that variable is excluded from the model. This increases the runtime significantly on large datasets
Call:
randomForest(formula = medv ~ ., data = Boston, mtry = 12, importance = T, subset = train.boston)
Type of random forest: regression
Number of trees: 500
No. of variables tried at each split: 12
Mean of squared residuals: 11.40162
% Var explained: 85.17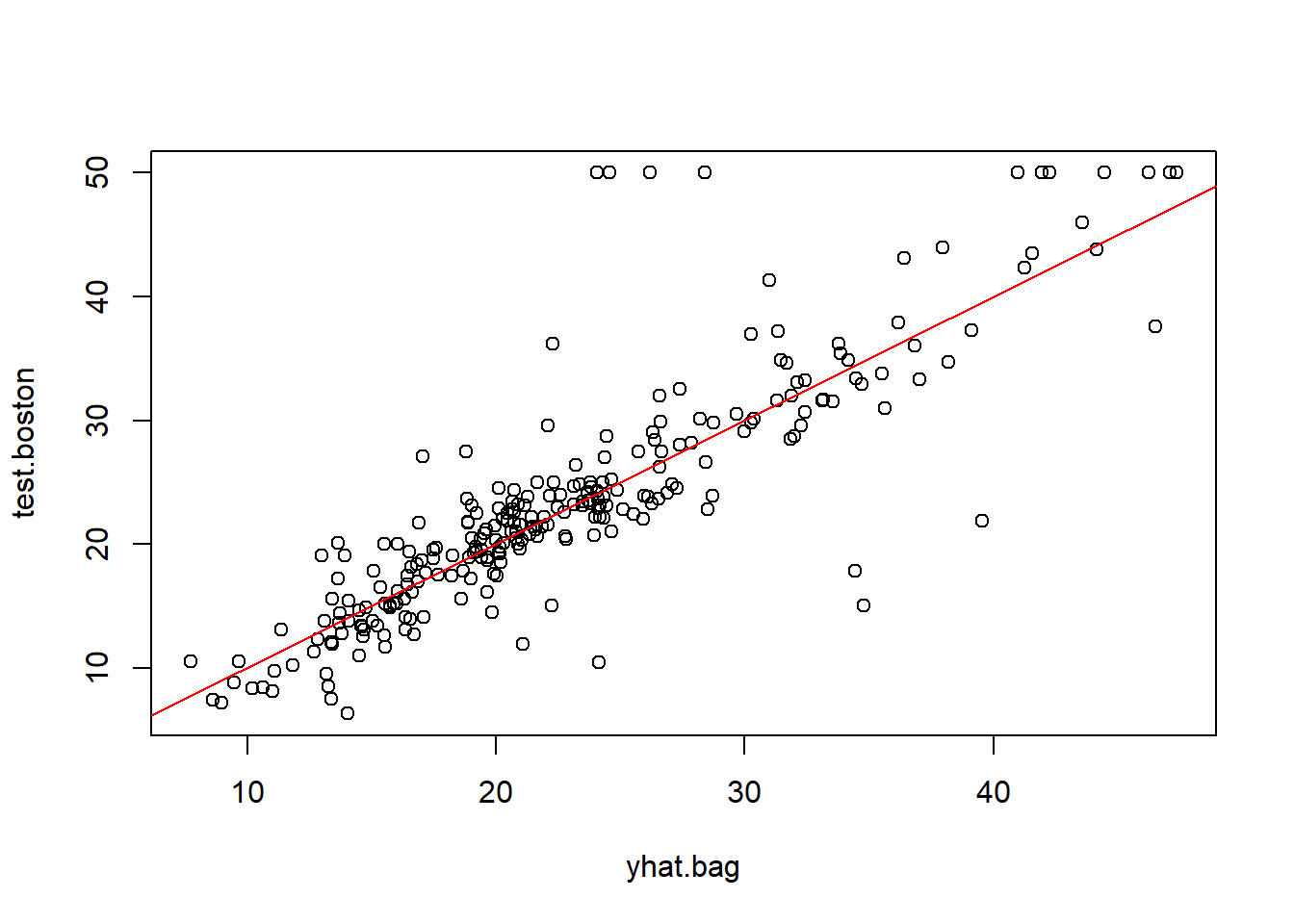
What’s the accuracy here? Checking the MSE
And square root of MSE or RMSE is:
That’s $4839 which is better than $ 5940 derived from the 7-node decision tree discussed in Key Takeaways. Moving to Random Forest now.
Its the same code, but we alter the number of predicted variables to mtry parameter
Default settings for randomForest()
for regression analysis,
for classification analysis,
Call:
randomForest(formula = medv ~ ., data = Boston, mtry = 6, importance = T, subset = train.boston)
Type of random forest: regression
Number of trees: 500
No. of variables tried at each split: 6
Mean of squared residuals: 10.09466
% Var explained: 86.87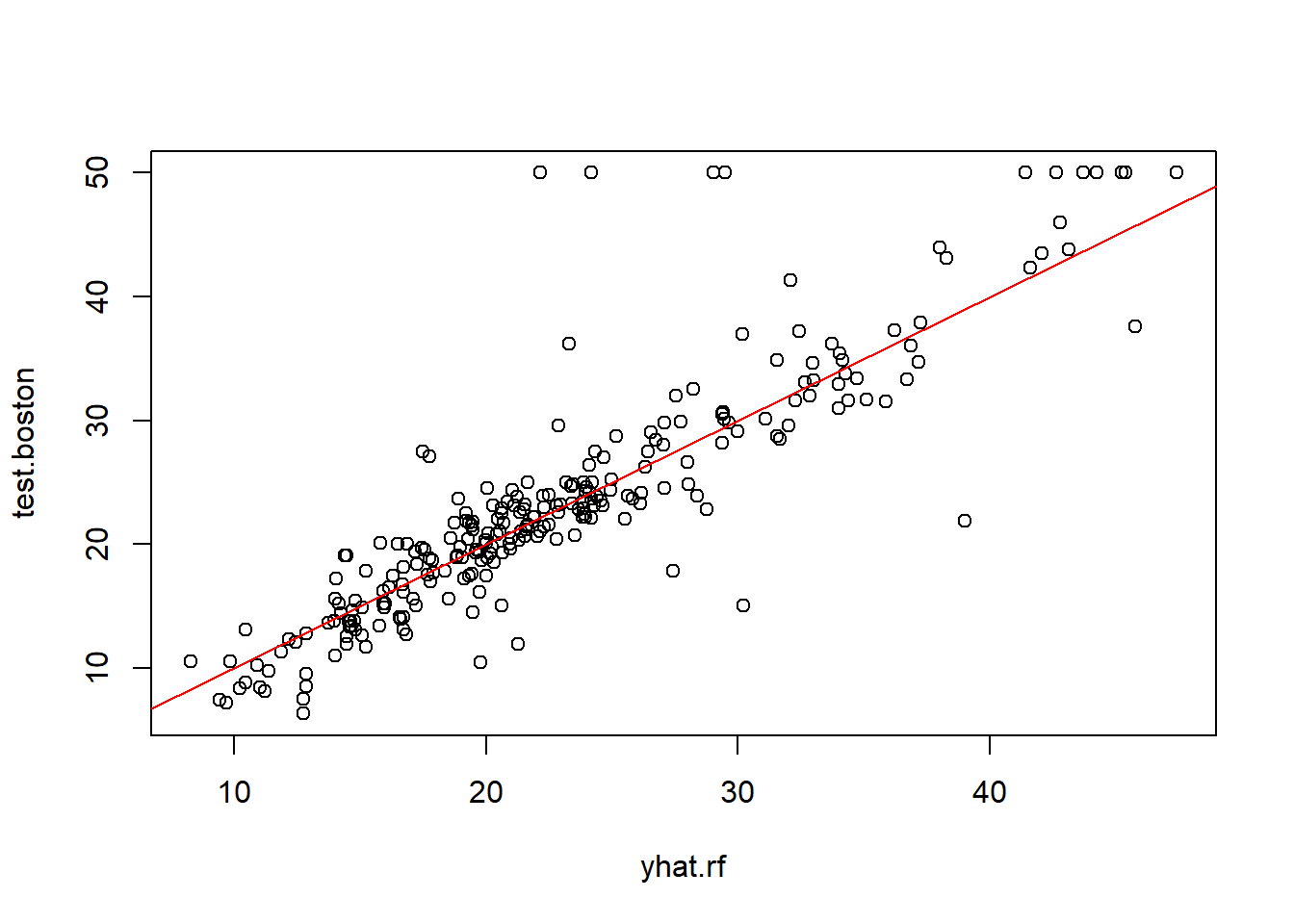
What’s the MSE here?
.. and therefore RMSE is:
That’s ±$4480 from the mean predicted values - which is better than $4839 by using the Bagging method.
Before moving ahead, we can also check the importance() function to determine key predictors
%IncMSE IncNodePurity
crim 19.435587 1070.42307
zn 3.091630 82.19257
indus 6.140529 590.09536
chas 1.370310 36.70356
nox 13.263466 859.97091
rm 35.094741 8270.33906
age 15.144821 634.31220
dis 9.163776 684.87953
rad 4.793720 83.18719
tax 4.410714 292.20949
ptratio 8.612780 902.20190
lstat 28.725343 5813.04833What are these columns?
in regression trees, the node impurity measured by the training Residual Sum of Squares(RSS)
in classification trees, it is the deviance
This shows that the two most important variables are rm (average number of rooms per dwelling) and lstat (lower status of the population in %)
Using the gbm package (Gradient Boosting Model) for boosted trees. Few notes:
distribution = "gaussian" is considered for regression trees. For classification, it should be distribution = "bernoulli"n.trees = 5000 is the number of trees we want to iterate overinteraction.depth = 4 limits the depth of each tree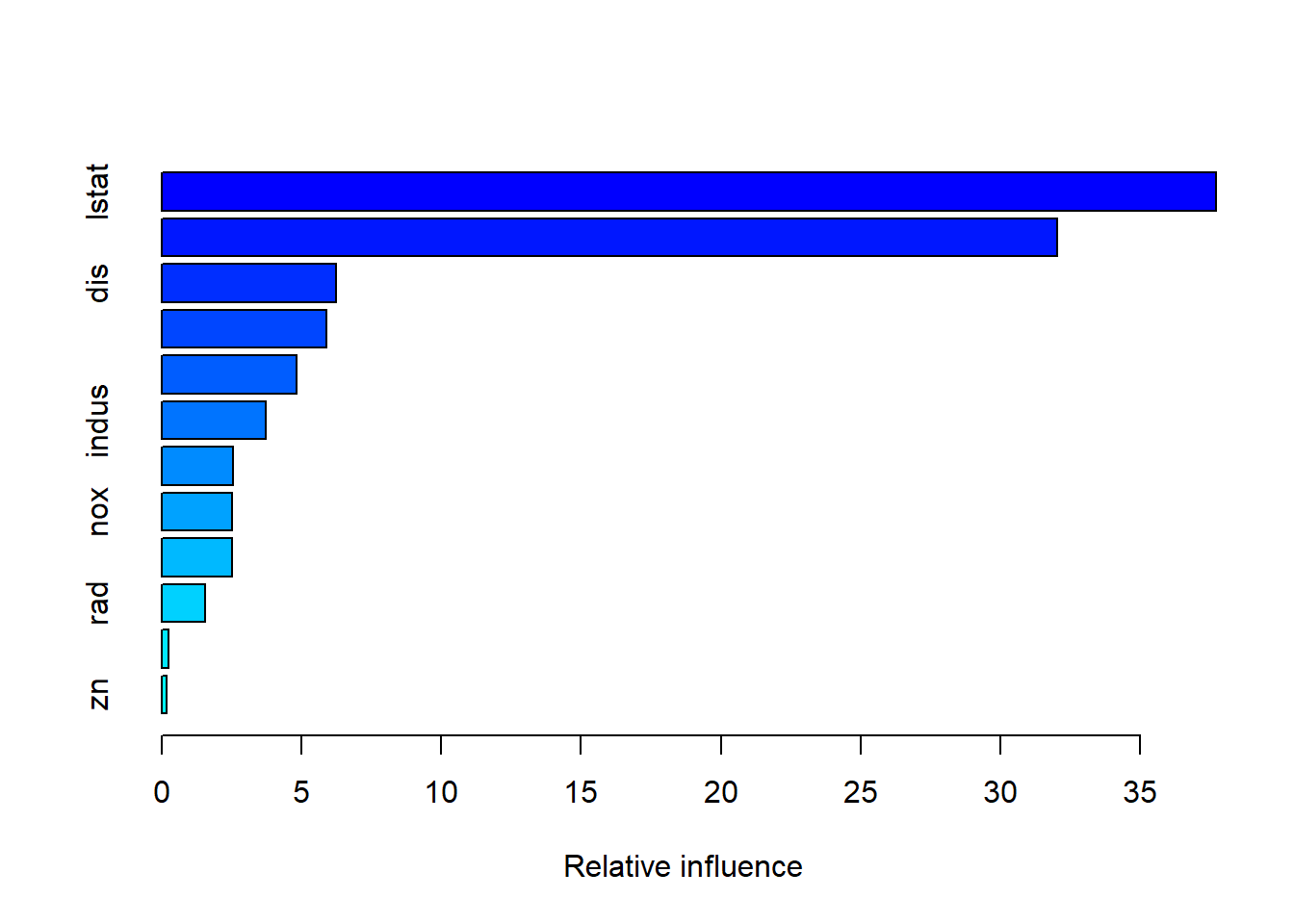
var rel.inf
lstat lstat 37.7145639
rm rm 32.0396810
dis dis 6.2532723
crim crim 5.9078403
age age 4.8163355
indus indus 3.7365846
tax tax 2.5457121
nox nox 2.5286998
ptratio ptratio 2.5091014
rad rad 1.5427771
chas chas 0.2451445
zn zn 0.1602876As seen earlier, lm and rstat show up as the most important variables.
By plotting the partial dependence of rm and lstat on outcome variable, we see that
rm has a direct relation viz. more the number of rooms, higher the price increaseslstat has an inverse relation viz. higher the lower stata in the neighbourhood, lower the price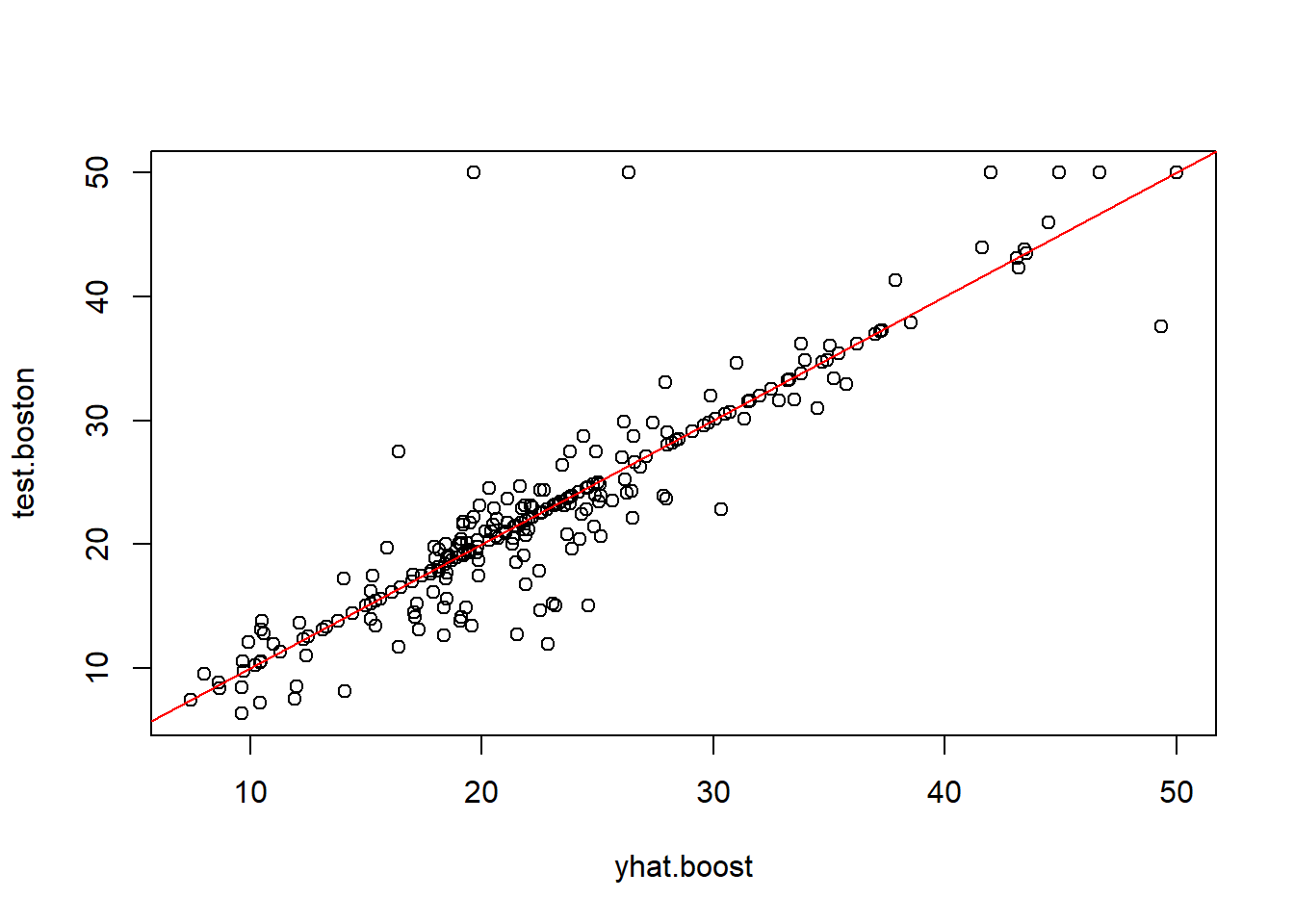
Figure looks so much better. testing the accuracy now. starting with the MSE
Wow.. that’s significantly lower. How about the RMSE?
Amazing. This means our predicted value on an average is ±$3603 from the actual which is a signifcant improvement from the RMSE calculated by Random Forest ±$4480
Also referred as the shrinkage parameter, the default value is 0.001 but we will change this to 0.01
The resulting MSE therefore is calculated as:
Now we’ve got it even lower at ±$3593
The function gbart() is used for regression analysis. This syntax slightly reminds me of the python syntax as we’re back to creating matrices for each test, train, x & y.
Creating the model now:
*****Calling gbart: type=1
*****Data:
data:n,p,np: 253, 12, 253
y1,yn: 0.213439, -5.486561
x1,x[n*p]: 0.109590, 20.080000
xp1,xp[np*p]: 0.027310, 7.880000
*****Number of Trees: 200
*****Number of Cut Points: 100 ... 100
*****burn,nd,thin: 100,1000,1
*****Prior:beta,alpha,tau,nu,lambda,offset: 2,0.95,0.795495,3,3.71636,21.7866
*****sigma: 4.367914
*****w (weights): 1.000000 ... 1.000000
*****Dirichlet:sparse,theta,omega,a,b,rho,augment: 0,0,1,0.5,1,12,0
*****printevery: 100
MCMC
done 0 (out of 1100)
done 100 (out of 1100)
done 200 (out of 1100)
done 300 (out of 1100)
done 400 (out of 1100)
done 500 (out of 1100)
done 600 (out of 1100)
done 700 (out of 1100)
done 800 (out of 1100)
done 900 (out of 1100)
done 1000 (out of 1100)
time: 9s
trcnt,tecnt: 1000,1000Computing the test error MSE
uhoh.. it was 12.9 for the Boosted RF trees. So the RMSE can be calculated as:
That’s ±$3993 which is not as good as $3593 RMSE that the Boosted Tree with shrinkage gave us.
The calculations show that as per the RMSE, the accuracy of models can be ordered as:
Bagging < Random Forest < BART < Boosting < Boosting w/ Regularization