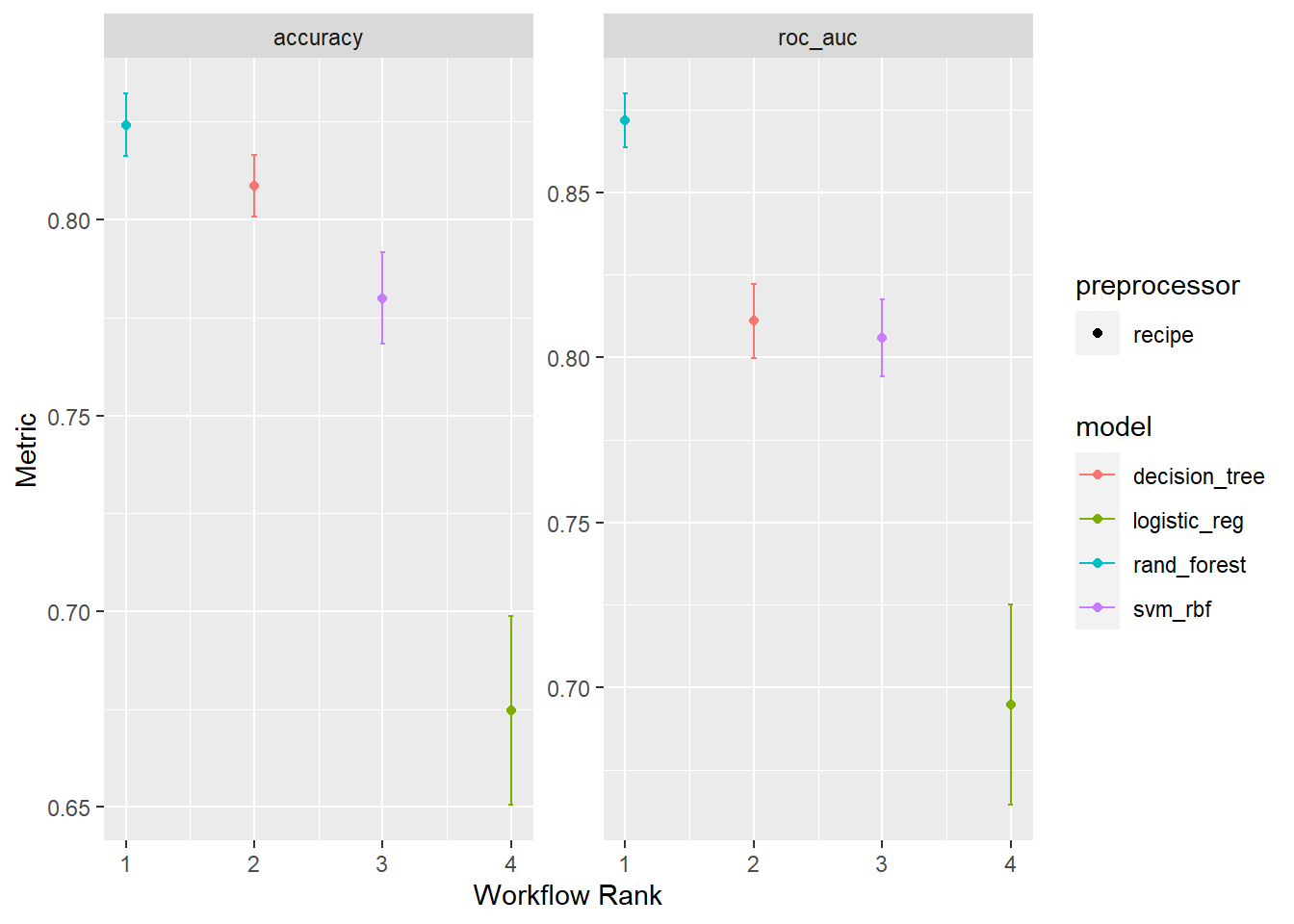
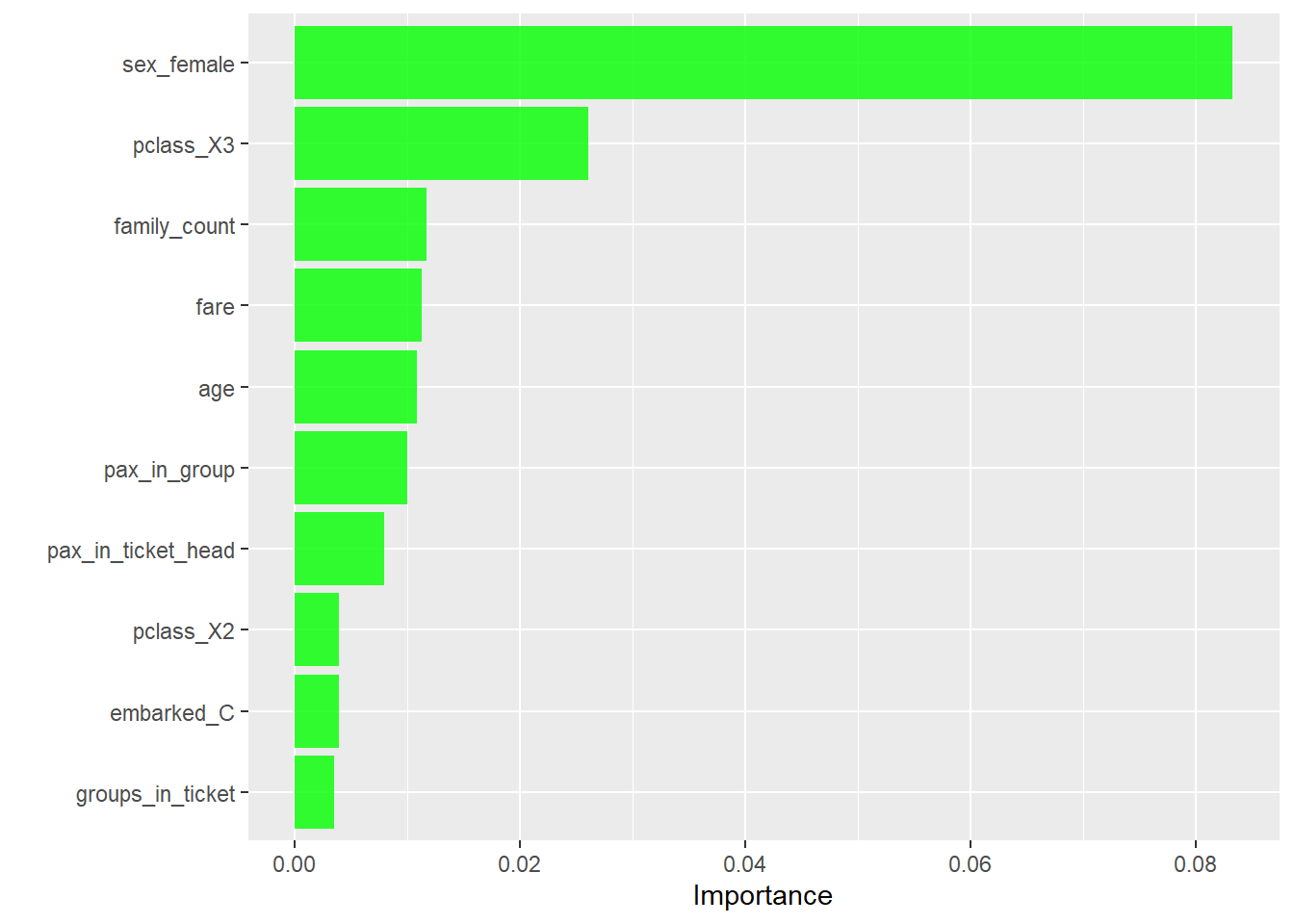
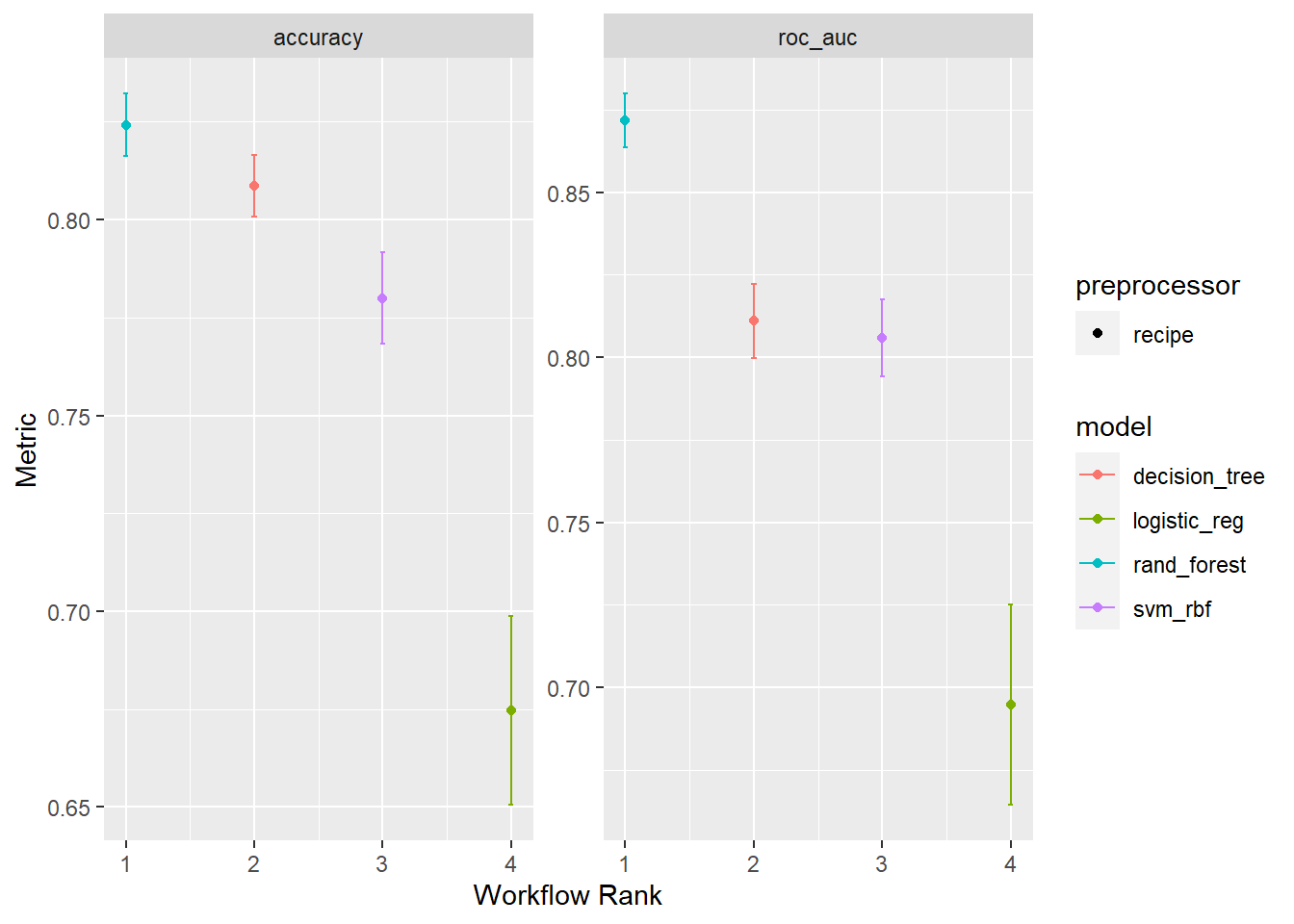
Rows: 1,309
Columns: 13
$ source <chr> "train", "train", "train", "train", "train", "train…
$ passenger_id <int> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, …
$ survived <fct> 0, 1, 1, 1, 0, 0, 0, 0, 1, 1, 1, 1, 0, 0, 0, 1, 0, …
$ sex <fct> male, female, female, female, male, male, male, mal…
$ age <dbl> 22, 38, 26, 35, 35, 22, 54, 2, 27, 14, 4, 58, 20, 3…
$ fare <dbl> 22, 38, 26, 35, 35, 22, 54, 2, 27, 14, 4, 58, 20, 3…
$ pclass <fct> 3, 1, 3, 1, 3, 3, 1, 3, 3, 2, 3, 1, 3, 3, 3, 2, 3, …
$ embarked <fct> S, C, S, S, S, Q, S, S, S, C, S, S, S, S, S, S, Q, …
$ family_count <dbl> 2, 2, 1, 2, 1, 1, 1, 5, 3, 2, 3, 1, 1, 7, 1, 1, 6, …
$ pax_in_group <int> 1, 2, 1, 2, 1, 1, 1, 5, 3, 2, 3, 1, 1, 7, 1, 1, 6, …
$ pax_in_ticket_head <int> 5, 14, 2, 8, 1, 1, 6, 5, 4, 4, 3, 14, 1, 22, 8, 1, …
$ groups_in_ticket <int> 1, 1, 1, 1, 1, 1, 2, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, …
$ final_grouping <fct> Braund_A/5 2117, Cumings_PC 1759, Heikkinen_STON/O2…